1. Database overview
1.1 Database overview
In plants, as most common RNA methylations, N6-methyladenosine (m6 A) and 5-methylcytosine (m5 C) play important dynamic roles in many biological processes, including embryonic development, stem cell fate determination, trichome branching, leaf morphogenesis, floral transformation, stress response, fruit maturation and root development. m6 A modification plays a vital role in almost every aspect of mRNA metabolism, including alternative splicing, 3’-end processing, nucleocytoplasmic shuttling, translation, and mRNA decay.
Cucume is mainly dedicated to the collection and standardization of RNA methylation sites in Cucurbitaceae. The methylation site data so far comes from our own experimental results: MeRIP-seq on cucumber and pumpkin. A large number of m5 C and m6A RNA methylation sites have been reported for the first time. In addition to adult plants, we use vascular exudation as samples for MeRIP-seq. We hope that this information will promote the study of vascular-mediated systemic communication, abiotic stress and disease resistance in Cucurbitaceae. In fact, we previously reported that a cucumber and pumpkin heterograft mobile RNA data set was integrated. Other published data: Cucumber QTL, GWAS SNP datasets are added to provide a basis for in-depth research on the epigenetics of Cucurbitaceae.
1.2 Datasets introduction
Cucumber and pumpkin grafting mobile RNAs
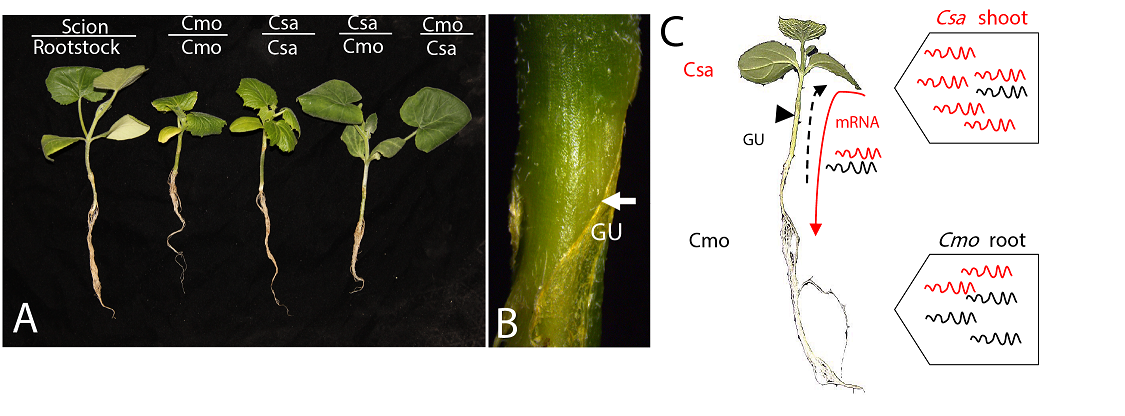
Hypocotyl grafting of Cucumis sativus and Cucurbitea moschata seedlings (Liu et al. , 2020). (A) Cucumis sativus (scion)/Cucurbitea moschata (rootstock) (Csa/Cmo) and reverse grafting (Cmo/Csa) heterograft combinations and homografts (Csa/Csa, Cmo/Cmo). (B) Graft-union magnification of Csa/Cmo shown in (A); arrowheads indicate graft union (GU). (C). Scion-to-rootstock long-distance movement of heterologous mRNAs through the phloem was identified in hypocotyl-grafted cucurbits. If Csa transcripts (red) moved to Cmo stock across the grafting union, they could be distinguished from Cmo transcripts (black) based on genomic single nucleotide polymorphism (SNP) differences. The first true leaf and whole root were sampled for further analyses. Dotted arrow indicates rootstock-to-scion mobile Cmo transcripts (black) moving to Csa through cell-to-cell spread or a cycle pathway of scion-to-rootstock-to-scion.
Cucumber quantitative trait locus (QTL)
The datasets of QTL regions were obtained from publicated literature, which contains 287 cucumber QTL for 31 quantitative traits. These traits are including QTL for whole plant vegetative growth and development, reproductive development, fruit–related traits, seed-related traits and disease resistances and abiotic stress tolerances. QTL mapping studies for these traits were conducted using multiple recombinant inbred lines populations such as the following crosses: 9110Gt × 9930, Gy14 × B10 and PI 183967 × 931. Through database integration of QTL interval information and RNA methylation sequencing results, the intensity of methylation distribution can be analyzed, and the potential association with agronomic traits can be explored.
Cucumber GWAS SNP
SNP associated with certain agronomical traits were obtained from published references (Liu et al. ,2020; Wang et al. ,2018). For possible connections, the GWAS SNP locates in methylated mRNAs can be queried.
1.3 Reference
1. Thieme, C. J., Rojas-Triana, M., Stecyk, E., Schudoma, C., Zhang, W., Yang, L., et al. (2015). Endogenous Arabidopsis messenger RNAs transported to distant tissues. Nat. Plants. 1:15025. doi: 10.1038/nplants.2015.25
2. Yang, L., Perrera, V., Saplaoura, E., Apelt, F., Bahin, M., Kramdi, A., et al. (2019). m 5 C methylation guides systemic transport of messenger RNA over graft junctions in plants. Curr. Biol. 29:2465. doi: 10.1016/j.cub.2019.06. 042
3. Zhang, W., Thieme, C. J., Kollwig, G., Apelt, F., Yang, L., Winter, N., et al. (2016). tRNA-related sequences trigger systemic mRNA transport in plants. Plant Cell. 28:1237. doi: 10.1105/tpc.15.01056
4. Liu, W., Xiang, C., Li, X., Wang, T., Lu, X., Liu, Z., et al. (2020). Identification of long-distance transmissible mRNA between scion and rootstock in cucurbit seedling heterografts. Int. J. Mol. Sci. 21:5253.
5. Wang, T., Li, X., Zhang, X., Wang, Q., Liu, W., Lu, X ., et al. (2021). RNA motifs and modification involve in RNA long-distance transport in plants. Frontiers in cell and developmental biology. 9: 651278.
6. Wang, Y., Bo, K., Gu, X., Pan, J., Li, Y., Chen, J ., et al. (2020). Molecularly tagged genes and quantitative trait loci in cucumber with recommendations for QTL nomenclature. Hortic Res. 7:3.
7. Liu, X., Lu, H., Liu, P., Miao, H., Bai, Y., Gu, X ., et al. (2020). Identification of Novel Loci and Candidate Genes for Cucumber Downy Mildew Resistance Using GWAS. Plants-Basel. 9:1659.
8. Wang, X., Bao, K., Reddy, U., Bai, Y., Hammar, S., Jiao, C., et al. (2018) The USDA cucumber (Cucumis sativus L.) collection: genetic diversity, population structure, genome-wide association studies, and core collection development. Hortic Res. 5:64
2. Database Usage
2.1 Homepage
In the homepage of the Cucume database, there are multiple modules for quickstart . We classify and organize the bioinformatics data related to methylation and present it to users in the form of tables or graphs. The function of these modules is to further optimize the appearance and layout of our website, simplify the operation logic, and provide a better experience for researchers in the field of RNA methylation.
2.2 Browse
To be neat and beautiful on the Browse homepage, the first display is the content of title 1. Users can check the items in 2. to get more information.
The information of m5C and m6 A comes from our MeRIP-seq experiment. The Adult plant sample is a mixture of cucumber or pumpkin roots, stems, and leaves, and the Vascular sample is vascular exudation. The numbers below them are the highest values of the corresponding methylation peaks. Mobile RNA is grafting mobile RNA. The number under TLS indicates the number of tRNA-like structures in the CDS region. TLS, mobile RNA, GWAS SNP, and QTL information all come from published information.

Click on a gene ID to enter the details page of the gene, such as the specific methylation site and GWAS SNP site.

2.3 Search
Users can search by ID or conditions.
The result will show the genes that meet the conditions, which is actually a subset of Browse. The displayed information and operation logic is the same.

2.4 QTL
To provide methylation-related genetic regulation and epigenetic research, we have added the QTL module. On the QTL homepage, currently only information about cucumber QTL (Cs QTL) is provided. After selecting the traits of interest at the top, the QTL corresponding to each trait is displayed in the picture. Users can click the corresponding position on the picture to enter the QTL details.
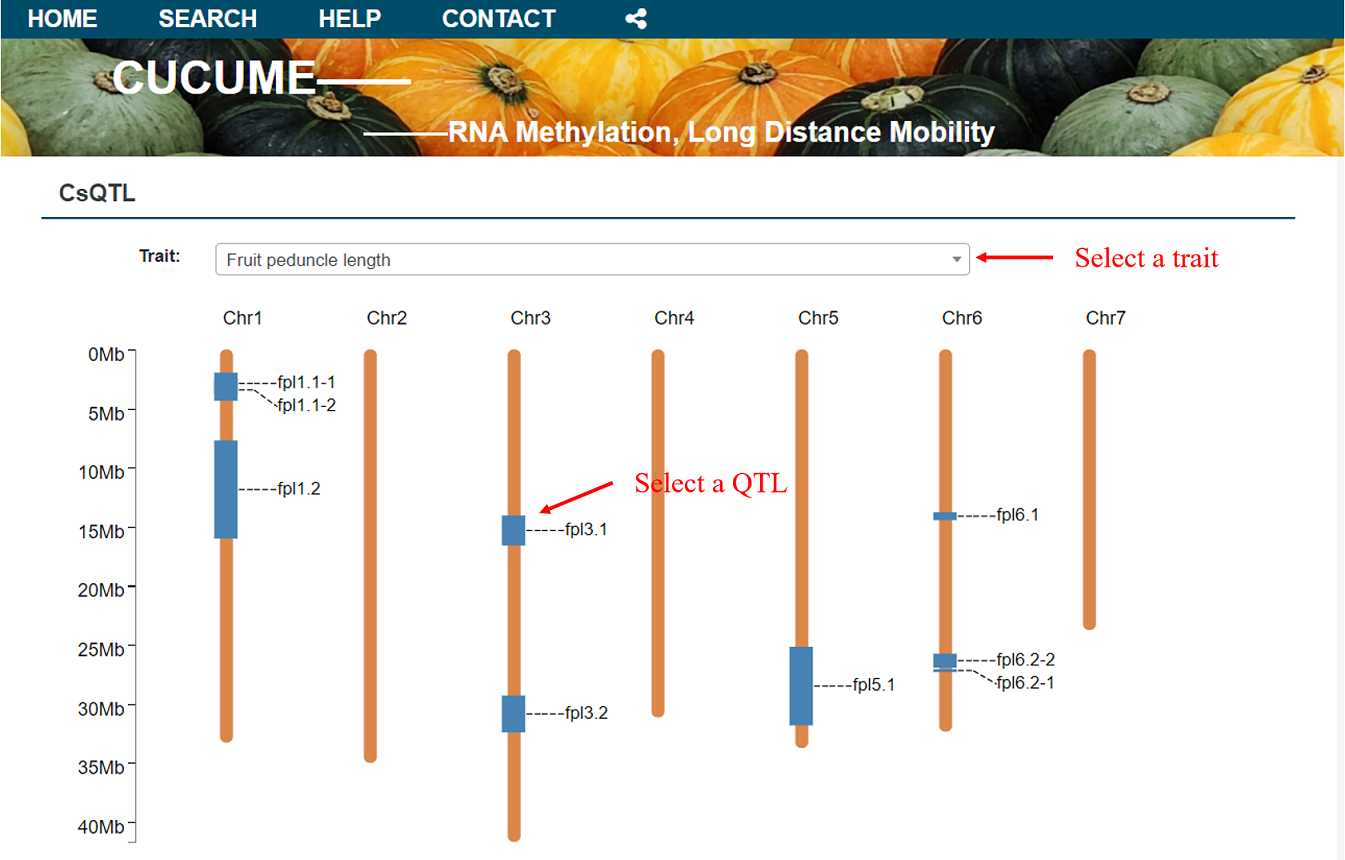
Alternatively the users can click on the QTL ID in the table below to enter the QTL details.
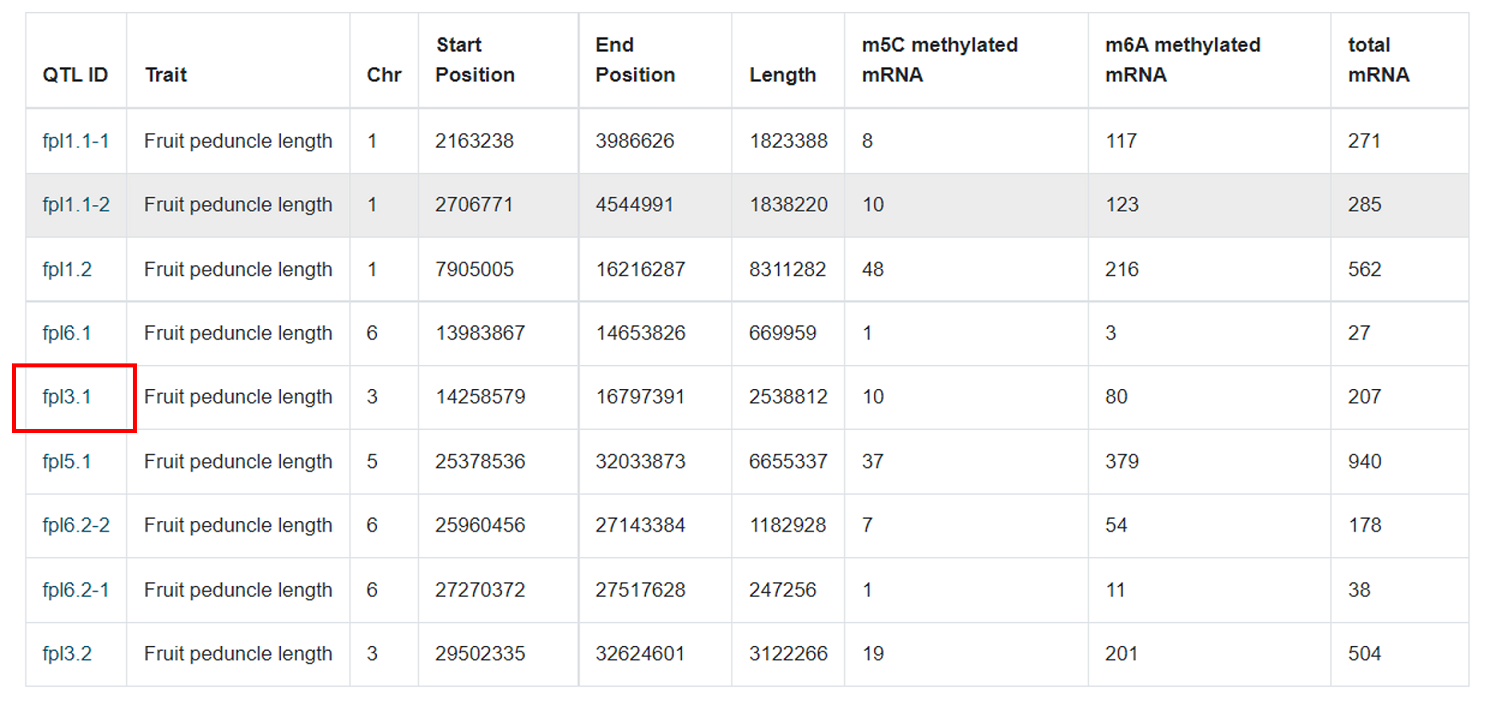
The details page provides basic information about the QTL and its m 6A or m5C methylation sites.
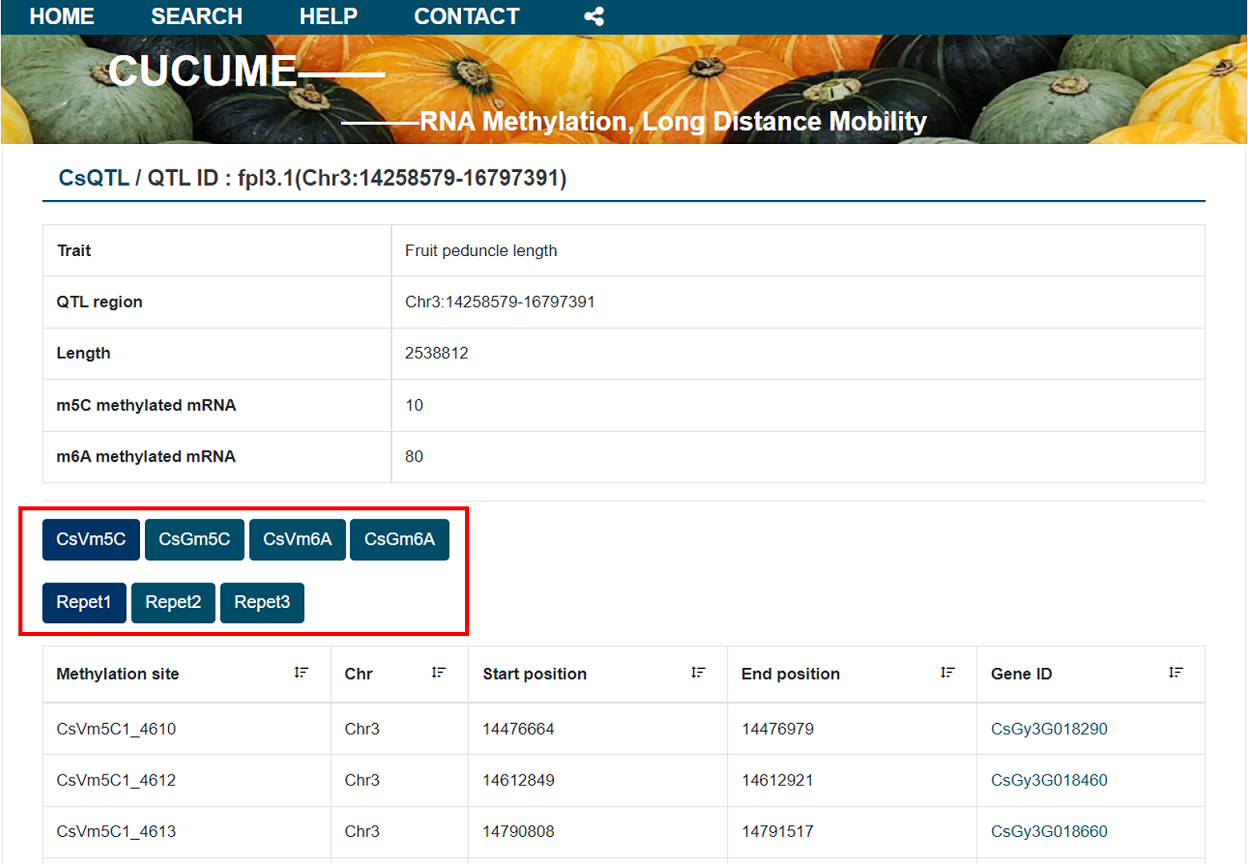
In this module, we have made preliminary statistics on the issues that users may be interested in.

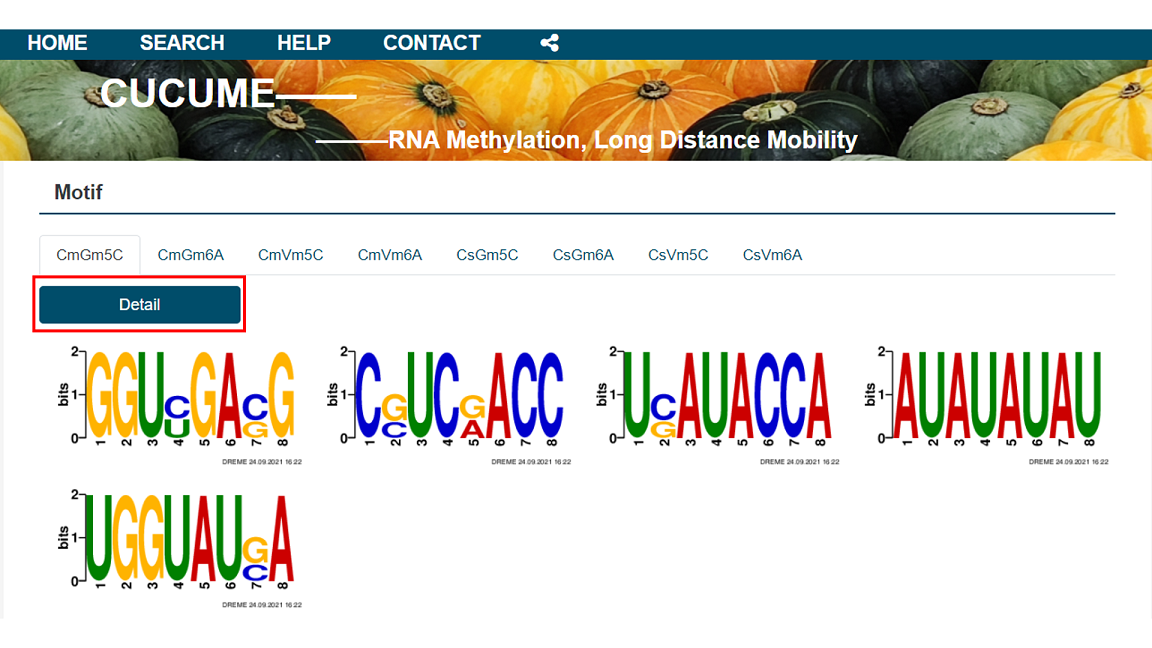
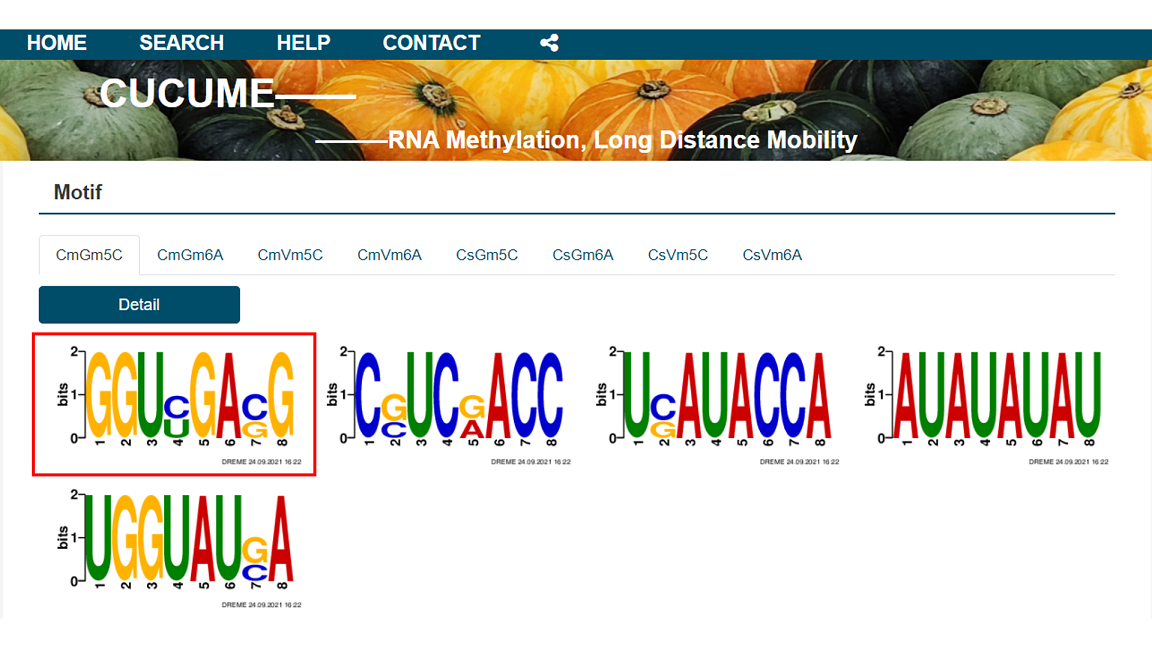
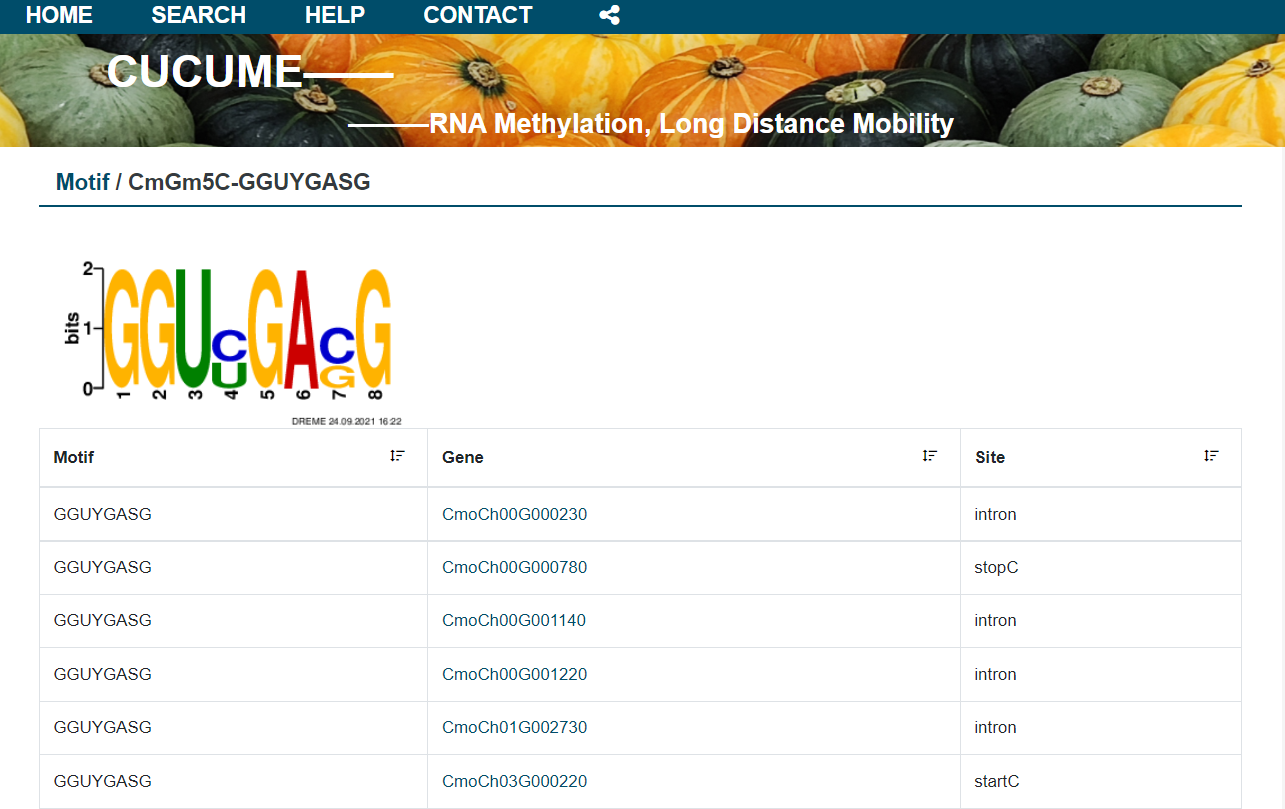
2.5 Motif
In the Motif module, the results of the search using the DREME program are presented per biological repetitions. The details of the analysis and the RNA contained in each motif can be provided for query.